Structural studies of membrane transporters
Most of our projects start with structural studies. In the past, they would require protein crystallization and the use of X-ray scattering to reconstruct the protein electron density and build a molecular model. Today, we conduct most of our structural studies using Cryo-EM that has dramatically facilitated the efforts. The first near-atomic resolution structures provide blueprints of the proteins, which allow educated guesses on their molecular mechanisms. But to fully dissect the mechanics of the transporters, multiple structural snapshots taken along their transport cycle are needed. Using these, we gain a more realistic mechanistic understanding. The development of Cryo-EM approaches facilitated structural determination under progressively more physiologic conditions. For example, we can determine structures of the transporters reconstituted into lipidic nanodiscs. In this way, we can directly visualize how the transporters interact with and sometimes perturb their membrane environment. Crystallography also has its unique advantages, allowing, for example, the use of anomalous diffraction to locate ion binding sites in some cases.
Methods to study transporter dynamics
The physiologic role of the membrane transporters is to carry their substrates across the cell membranes. Different transporters use different mechanisms to do the job. Still, they share the critical mechanistic principle, where they are providing so-called alternating access to the substrate-binding site from the opposite sides of the membrane. This process invariably involves at least one global structural rearrangement of the transporter. For example, glutamate transporters function by an “elevator”-like mechanism, where the substrate- binding domain moves from the extracellular to the intracellular side of the membrane and back. The dynamics of the structural transitions determine the rates of transport and therefore are integral to their function. We have been interested in developing new methods to follow these dynamics, mostly in collaborations with other labs. For example, recently, in partnership with David Eliezer’s lab, we have developed a 19F NMR approach to detect multiple conformational states accessed by the transporter. We showed how to relate them to known molecular structures and estimate the rates of the conformational exchange.
Transporters dynamics and function, one molecule at a time
With long-term help from Scott Blanchard’s group, we have established in the lab single-molecule FRET microscopy to follow structural dynamics in individual transport molecules. You can learn a lot about how birds fly by observing one of them at the time, rather than watching a whole flock at once. By the same token, a wealth of dynamic information is accessed through single-molecule methods following the motions of individual proteins. We couple these studies of the transporter dynamics with new approaches to follow the transport activity of single transporters.
Developing transporter activators
Many drugs target membrane transporters and most of them serve as inhibitors. But sometimes, it is desirable to have drugs that work as activators of the transporters. However, when we think about the working cycle of a transporter, it is not clear what a small molecule could do to speed it up. We are interested in conceptual development of activators and also in understanding the mechanisms of the existing activators.
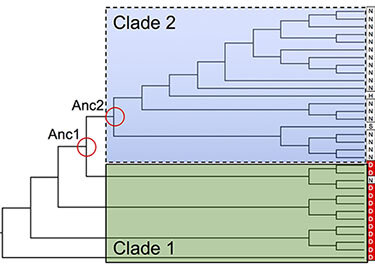
Evolution of transporters
Over millennia, the transporters evolve to address the physiological needs of the organisms. Some even develop novel functions that were absent in their ancestors. Sometimes, analysis of the extant sequences shows apparent differences between transporters with different functions. Other times, however, these differences are obscure and difficult to pinpoint. We are interested in understanding what are the evolutionary mechanisms behind such subtle molecular differentiations.